


2.6 Evidence from Molecular Biology and Genetics
2.6.1 DNA as Genetic Material. As was stated earlier (see
I.1.4), genetic variability arising from mutation and recombination
by natural selection serves as the raw material of evolution. Before the
advent of molecular biology, very little was known concerning the nature
and mode of action of the gene. Later, the identification of biological
macromolecules in cells prompted many to explore the relationships between
these substances and the Mendelian concepts (see I.1.3) of particulate
genes. [145]
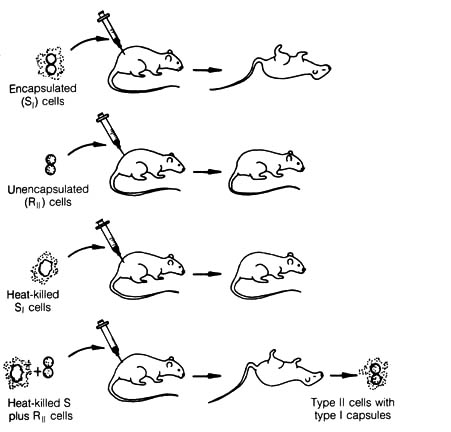
Figure 2.41. Bacterial Transformation. Transformation
was discovered by F. Griffith, who noted that encapsulated (smooth or S)
diplococci
cause a fatal infection in mice, whereas nonencapsulated (rough or R) cells
do not. Heat-killed S cells are likewise harmless, except when mixed with
live R cells. In the latter case, a fatal infection can occur, and live
cells having capsules characteristic of the S strain are found. Reprinted,
with permission, from Dyson, R. D. Cell biology, a molecular approach.
1st ed. Boston: Allyn and Bacon, Inc.; 1974.

Figure 2.42. The Transforming Principle. When encapsulated
diplococci
were chemically fractionated, and the transformation experiment performed
in vitro with each fraction one at a time, O. T. Avery and his colleagues
found that DNA is the agent responsible for transformation. Reprinted,
with permission, from Dyson, R. D. Cell biology, a molecular approach.
1st ed. Boston: Allyn and Bacon, Inc.; 1974.
The first experiment that paved the way for understanding the chemical
[146] nature of the gene was the discovery of transformation in
the bacteria Diplococcus pneumoniae by F. Griffith in 1928 (1).
The pathogenicity of the D. pneumoniae bacterium is associated with
the existence of a capsule. The capsule is a slimy material that
surrounds the cell and prevents phagocytosis by the white blood cells of
the host organisms. A nonpathogenic strain lacks the capsule and is easily
destroyed by host white blood cells. Griffith demonstrated that there were
stable materials from the heat-killed pathogenic strain of D. pneumoniae
(S
form) that could be transmitted and incorporated into the nonpathogenic
strain (R form). The materials could induce the latter to synthesize the
capsule and thereby transform the bacterium from a nonpathogenic to a pathogenic
strain (Figure 2.41). In 1944, Avery, Macleod, and McCarty (2)
identified the material responsible for this genetic transformation of
D.
pneumoniae
as DNA (Figure 2.42).
With the establishment of the chemical nature of the gene, the next
important questions asked were (1) What is the chemical structure of DNA?
and (2) How are genetic messages encoded in DNA? The answers to these
two questions were provided by J. Watson and F. Crick (3,
4)
and made a great impact on modern biology. The answers opened up the new
discipline of molecular biology that presently is advancing rapidly and
is influencing every area of modern biological thinking.
For a chemical model to account for the functions of a gene, it must
be able to show two characteristics: (1) the potential to duplicate itself
with exact fidelity so that each of the daughter molecules will be a replica
of itself and (2) the ability to carry coded information that specifies
a particular set of traits characteristic of a certain line of descent.
Watson and [147] Crick postulated a double helix model of DNA that satisfies
both of these criteria (Figure 2.43).
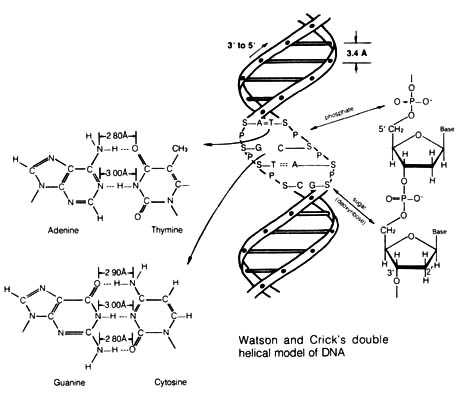
Figure 2.43. Base Pairing in DNA. A pyrimidine nucleotide
(cytidine or thymidine) is always paired with a purine nucleotide (adenosine
or guanosine), thus maintaining uniform overall dimensions in each pair.
The hydrogen-bonding capabilities of the common form of the bases lead
specifically to 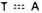 and
and  pairing. Reprinted,
with permission, from Dyson, R. D. Cell biology, a molecular approach.
1st ed. Boston: Allyn and Bacon, Inc.; 1974.
pairing. Reprinted,
with permission, from Dyson, R. D. Cell biology, a molecular approach.
1st ed. Boston: Allyn and Bacon, Inc.; 1974.
The structure of the double helix (DNA) can be viewed from several levels.
The first and most important level is the bases, namely, the pyrimidines
thymine and cytosine, and the purines guanine and adenine (Figure 2.44).
They can form metastable paired configurations by hydrogen bonding (i.e.,
adenine paired with thymine via two hydrogen bonds; guanine paired with
cytosine via three hydrogen bonds) (see Figure 2.43). The second
level is the nucleoside. This is the combination of a single base
with a 5 carbon (pentose) sugar having a 5 membered furanose ring. The
absence of an oxygen at the 2' carbon position of the ring makes the sugar
of the nucleoside deoxyribose. At the third level, there are nucleotides
consisting
of a nucleoside with an added phosphate group at the 5' position of the
sugar. The fourth level involves the joining together of nucleotides by
hooking up the 5' phosphate group of one with the 3' [148] position on
the sugar ring of another, producing a long chain of nucleotides called
a polynucleotides (Figure 2.45). The fifth and highest level involves two
adjacent polynucleotide chains bound together by a long series of hydrogen
bonds between the complementary bases forming a double helical structure.
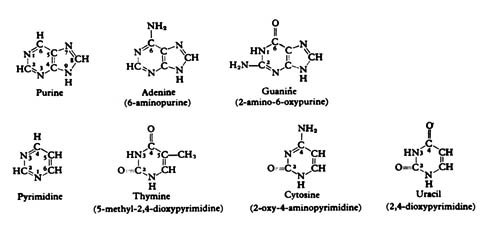
Figure 2.44. Purines and pyrimidines. Reprinted, with
permission, from Dyson, R. D. Cell biology, a molecular approach.
1st ed. Boston: Allyn and Bacon, Inc.; 1974.
The double helical structure of DNA can readily account for replication.
As soon as the double helix unwinds and separates, each strand serves as
a template for the synthesis of the missing strand. Because of the specificity
imposed on the pattern of hydrogen bonding between the adenine-thymine
pair and the guanine-cytosine pair, DNA replication proceeds with high
fidelity. During replication the parent strands of the DNA helix separate
and a complementary daughter strand is synthesized on each parent strand.
Thus two DNA molecules identical to the original molecule are produced
(Figure 2.46).
2.6.2 Gene Expression and the Genetic Code. The action of the
genetic material in an organism can be summarized in the central dogma
of molecular biology: DNA -> RNA -> protein. The information in the DNA
is transcribed into the messenger RNA that is then translated into protein
(Figure 2.47).
The RNA's or ribonucleic acid transcribed from the DNA is single stranded
and contains the bases adenine, guanine, and cytosine as in DNA, but a
new base uracil substitutes for thymine (Figure 2.44; 2.47). Also RNA has
an oxygen at the 2' position of each sugar ring (see Figure 2.45)
that is not present in DNA. Each different RNA is transcribed on only one
of the DNA strands by complementary pairing facilitated by hydrogen bonding
between bases (Figure 2.47a).
There are three types of RNA that can be transcribed from the DNA: messenger
RNA, transfer RNA, and ribosomal RNA. Each transfer RNA carrying an amino
acid makes contact with a messenger RNA at the ribosome [149] some through
base pairing of a triplet base sequence (colon)
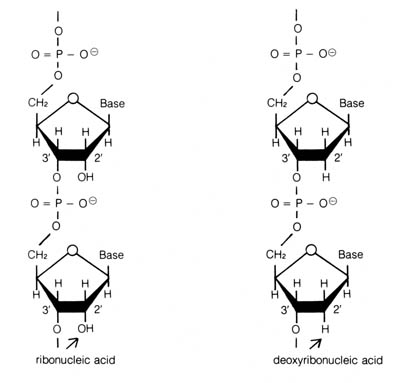
Figure 2.45. Presence of an oxygen in the 2' position
of each sugar ring of the ribonucleic acid as contrasted with deoxyribonucleic
acid.
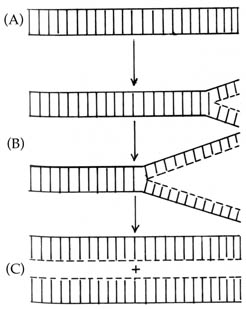
Figure 2.46. DNA replication. (A) Intact DNA double
helix. The two horizontal lines represent the sugar phosphate backbone.
The vertical lines represent hydrogen bonding between complimentary bases
(see Figure 2.43). (B) DNA unwinds and separates. New DNA (---)
is synthesized using the unwound strand of the parental DNA as a template.
(C) Two daughter DNA double helices with one strand contributed by the
parental molecule and one newly synthesized strand.
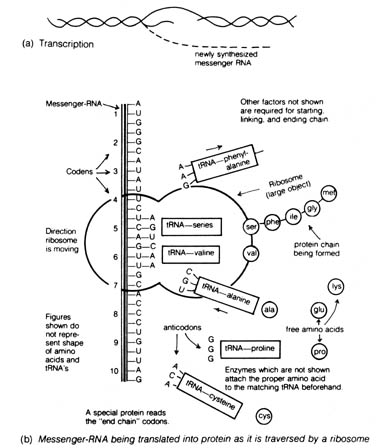
Figure 2.47. Transcription and translation. [150] As each
colon or triplet of letters is read, a transfer-RNA molecule approaches
which has an anticodon that will base-pair with those letters. This tRNA
carries its matching amino acid which has been attached to it by its interpreter
enzyme. As the tRNA is processed by the ribosome, the amino acid is joined
onto the forming protein chain in the order called for by the mRNA sequence
of code letters, which in turn was transcribed shortly before from the
DNA master copy. This complex process takes place with fantastic speed
and precision, and is remarkably similar in all living things known, from
amebas to human beings. Recent evidence indicates that both transcription
and replication may often be associated with cell membranes, including
the endoplasmic reticulum. Adapted, with permission, from Coppedge,
J. F. Evolution: possible or impossible? Grand Rapids, MI: Zondervan
Publishing House; 1973.
of the messenger RNA and a complimentary sequence (anticodon) of the transfer
RNA (Figure 2.47b). Peptide bonds are formed between adjacent amino acids
at the ribosomal level. By a process of translocation or the movement [151]
of the ribosome along the messenger RNA, each successive codon of the messenger
RNA binds with another transfer RNA that loses its amino acid to the growing
amino acid polypeptide chain through peptide bond formation.
By the elegant experiments of Nirenberg and Mattaei (5);
Nishimura, Jones, and Khorana (6); and others, the genetic
code was deciphered. The 20 amino acids present in proteins (Table 2.15)
are specified by 64 triplet
code words in DNA. These code words
pass on the information via the codons in the messenger RNA and
the anticodons in the transfer RNA, thereby specifying the amino
acid sequence of the gene product. Since there are more code words than
amino acids, a phenomenon known as degeneracy was observed, i.e., each
amino acid is specified by more than one code word. Table 2.16 lists all
the possible combinations of the triplet colons on the messenger RNA and
the corresponding amino acids they specify.
There are also initiation and termination colons that
are involved in the punctuation of the genetic message. The initiation
colons GUG and AUG specify a particular amino acid named N-formylmethionine,
which starts every polypeptide chain. However, when GUG and AUG are found
in the middle of a polypeptide message, they specify instead valine and
methionine respectively. The termination or nonsense colons are the UAA,
UAG, and UGA colons that specify no amino acids. Thus the polypeptide chain
falls off the ribosome as soon as it reaches one of these colons. Delimitated
by the initiation and the termination codons, a gene is defined as the
number of consecutive triplet codes on the DNA that determines the primary
sequence of a polypeptide chain — the so-called one-gene, one-polypeptide
concept. This concept has been well documented in the genetic systems of
bacteria and bacterial viruses and has provided modern biologists with
the first operational tool to examine the interaction of genes.
The genetic code is probably universally applicable. The codons UUU,
AAA, and CCC have been found to specify phenylalanine, lysine, and proline,
respectively, in cell extracts prepared from a variety of different organisms
ranging from bacteria to mammals (7). Purified messenger
RNA encoding the protein hemoglobin extracted from rabbits was shown to
be able to direct the synthesis of rabbit hemoglobin in frog oocytes, suggesting
the colons in rabbits and frogs are translated the same way (8).
The apparent universality of the genetic code has been cited as evidence
that all living organisms arose from a single origin. After the early evolution
of the genetic code in organisms, it is thought to have remained constant
over a long period while other features of the organisms [152]
Table 2.15. The common amino acids.*
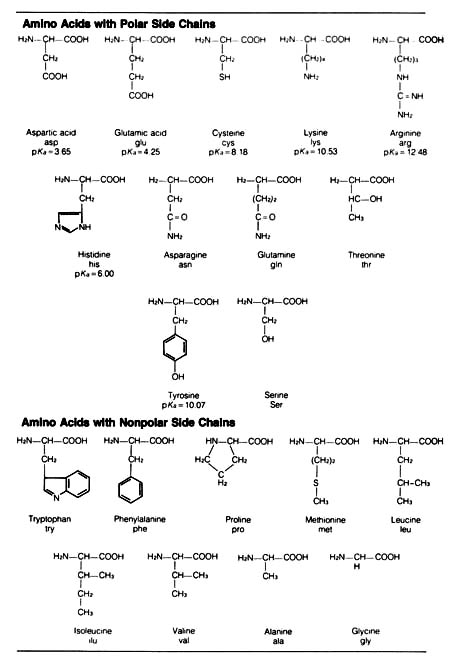
*The usual three-letter abbreviation is given, along with
the pKa of side chain groups that may carry significant charge at
physiological pH.
 NOTE:
Reproduced, with permission from Dyson, R. D. Cell biology, a molecular
approach. 1st ed. Boston: Allyn and Bacon. Inc.; 1974.
NOTE:
Reproduced, with permission from Dyson, R. D. Cell biology, a molecular
approach. 1st ed. Boston: Allyn and Bacon. Inc.; 1974.
diverged [153] in subsequent evolution. However, the constancy of
the evolution of the genetic code may be challenged if examples of discrepancy
are discovered in future research. The presence of odd guanosine triphosphate
at the 5' end and polyadenosine monophosphate at the 3' end of the eucaryotic
messenger RNA with obscure functions may suggest that there are new mechanisms
of the expression of the genetic code yet to be discovered (9).
Alternatively, the universality of the code could be attributed to a Creator's
master design that enables all living organisms to operate under a similar
set of physiological conditions.
Proteins are ubiquitous components in the cell. They serve as the building
blocks of cellular structure, and in the form of enzymes they also catalyze
chemical reactions in the metabolic pathways of the cell. The functions
of proteins are controlled by their structure that can be described at
four levels: primary, secondary, tertiary, and quarternary.
Table 2.16. The genetic code.*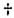

*Note that in all cases but two (Tyr, Met), the
third position may be occupied by either of the two purines or either of
the two pyrimidines without changing the coding specificity. The terminator
codons—UAA, UAG, and UGA—stop amino acid incorporation and free the growing
polypeptide chain. (The names ochre, amber, and opal refer to the mutant
bacterial strains in which the action of these terminators were first studied.)
Chain initiation begins with AUG or GUG, marked with an asterisk (*), either
of which can code (in procaryotes) for N-formylmethionine in addition
to the amino acid shown for it.
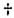 NOTE:
Reprinted, with permission, from Dyson, R. D. Cell biology, a molecular
approach. 1st ed. Boston: Allyn and Bacon, Inc.; 1974.
NOTE:
Reprinted, with permission, from Dyson, R. D. Cell biology, a molecular
approach. 1st ed. Boston: Allyn and Bacon, Inc.; 1974.
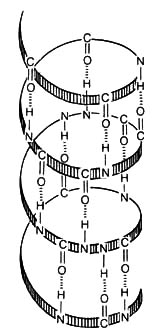
Figure 2.48. The alpha helix maintained by intrachain
hydrogen bonding. [154]
The primary structure is the linear sequence of amino acids that are
joined together by peptide bonds to form a polypeptide chain. There are
20 naturally occurring amino acids in the living world — 11 that are charged
ionic forms (those with polar side chains) and 9 that are neutral (those
with nonpolar side chains) under physiological conditions (Table 2.15).
Each species of protein has a unique amino acid sequence that essentially
determines its secondary, tertiary, and quarternary structure. One type
of secondary structure is the a helix configuration of a polypeptide chain
(Figure 2.48) (the envelope in Figure 2.49) formed by intrachain hydrogen
bonding. The tertiary structure is the apparent globular shape of the polypeptide
in solution when it coils together by intrachain, mostly noncovalent forces
between amino acid side chains. Proteins with more than one polypeptide
chain have quarternary structure, each chain being a subunit. The position
of each subunit is stabilized by noncovalent forces between chains (Figure
2.49). Hemoglobin (Figure 2.49), the oxygen-carrying protein in vertebrate
red blood cells, has a quarternary structure consisting of two 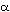 and two
and two 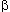 chains.
chains.
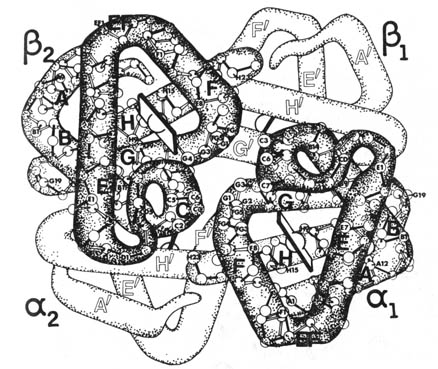
Figure 2.49. Hemoglobin. Reproduced from Dickerson and
Geis, slide set, Molecular structure of protein. By permission from Dr.
Richard E. Dickerson, Department of Chemistry, California Institute of
Technology, Pasadena.
The function of any protein depends on the nature of its three-dimensional
structure that is a reflection of the tertiary and quarternary [155] structure
state. This can best be understood by the lock-and-key theory of enzyme
function. The theory states that each enzyme (all enzymes are proteins)
has a specific configuration that fits the substrate (Figure 2.50). If
the functional three-dimensional structure of the enzyme is destroyed,
the reaction catalyzed by the enzyme does not take place.
Many inheritable phenotypic traits can be attributed to abnormal structure
of a single protein. The frequently cited example of a congenital disease
is sickle cell anemia (see I.3.2.l.c). This disease is caused by
the replacement of a single amino acid in the sixth position of the 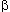 chain of hemoglobin. This replacement distorts the three-dimensional structure
of hemoglobin and causes the stacking of adjacent molecules in such a way
that the red blood cells carrying these distorted hemoglobins will become
sickle shaped and clump together in the absence of oxygen (Figure 2.51).
The sickle red blood cells last only half as long as normal cells, and
clumping of the sickle cells causes severe damage in vital organs. Therefore,
sickle cell anemic patients seldom live beyond 30 years of age. This [156]
has been called a molecular disease since the cause can be traced
to the distortion of a single molecule induced by a simple amino acid replacement.
chain of hemoglobin. This replacement distorts the three-dimensional structure
of hemoglobin and causes the stacking of adjacent molecules in such a way
that the red blood cells carrying these distorted hemoglobins will become
sickle shaped and clump together in the absence of oxygen (Figure 2.51).
The sickle red blood cells last only half as long as normal cells, and
clumping of the sickle cells causes severe damage in vital organs. Therefore,
sickle cell anemic patients seldom live beyond 30 years of age. This [156]
has been called a molecular disease since the cause can be traced
to the distortion of a single molecule induced by a simple amino acid replacement.
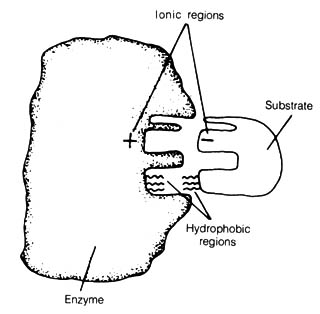
Figure 2.50. Enzyme-Substrate Interaction. According to
the lock-and-key theory, enzyme and substrate have complementary configurations.
In this diagram, matching spatial conformations permit ionic and hydrophobic
interactions to take place between the enzyme and its specific substrate.
Reprinted, with permission, from Dyson, R. D. Cell Biology, a molecular
approach. 1st ed. Boston: Allyn and Bacon, Inc.; 1974.

Figure 2.51. Photomicrograph of normal (dislike) and sickle
(crescent-shaped) blood cells. Reprinted, with permission, from Lehninger,
A. L. Biochemistry. 2nd ed. New York: Worth; 1975. Reprinted by
permission from Walter Dawn, National Audubon Society, New York.
The production of enzymatic proteins is known to be a delicately regulated
cellular process. The best-documented case of this type of gene expression
is that of the operon model, which has been demonstrated in bacterial
systems. An operon is a composite of several genes, usually clustering
together in the bacterial chromosome and involved in similar functions.
These genes are regulated by a single operator gene as the result of its
interaction with the product of a regulatory gene. [157]
The OPERON Model
OPERON: A single operator switches on a sequence of genes
and it and the structural genes it controls comprise an integrated unit
as both the physical and functional levels.
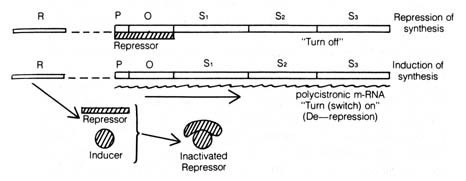
Figure 2.52. The Jacob-Monod operon model for control
of the synthesis of lactose-metabolizing enzymes. The repressor produced
by the regulator gene (R) normally binds to the operator gene (O)
and stops the transcription of the structural genes (S1, S2,
S3) by blocking the RNA polymerase from binding to the promoter
gene (P). In the presence of the inducer (lactose), the repressor
is bound by it, and a conformational change takes place that inactivates
the repressor from binding to the operator gene. Transcription of the structural
genes gives rise to polycistronic messenger RNA (m-RNA) that is a continuous
piece of RNA spanning across several genes. This m-RNA is then translated
to form the three enzymes required before lactose can be used as an energy
source.
The best exemplified system of the operon is the lactose operon of
the colon bacteria Escherichia coli (10) (Figure
2.52). The lactose operon is composed of three consecutive structural genes,
each coding for a different enzyme: 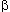 galactosidase that hydrolyzes the sugar lactose to galactose and glucose,
galactoside permease that enables the galactose sugar to enter the cell,
and galactoside acetylase that puts an acetyl group on the galactoside.
The expression of these structural genes depends on the action of three
other genes, namely, the regulator gene that synthesizes a protein repressor;
the operator gene to which the repressor protein binds; and the promotor
gene on which RNA polymerase (the enzyme responsible for the transcription
of the structural genes into RNA) binds and starts RNA synthesis.
galactosidase that hydrolyzes the sugar lactose to galactose and glucose,
galactoside permease that enables the galactose sugar to enter the cell,
and galactoside acetylase that puts an acetyl group on the galactoside.
The expression of these structural genes depends on the action of three
other genes, namely, the regulator gene that synthesizes a protein repressor;
the operator gene to which the repressor protein binds; and the promotor
gene on which RNA polymerase (the enzyme responsible for the transcription
of the structural genes into RNA) binds and starts RNA synthesis.
The intriguing feature of this model is that the structural genes are
expressed only when the operator gene is not bound by the repressor molecule.
This can be brought about when an inducer, namely, lactose, is present
and combines with the repressor to inactivate it. The enzymes encoded by
the structural genes can thereby be synthesized and the inducer metabolized.
As the lactose concentration decreases, more repressor [158] is freed to
interact with the operator gene again and shut off the transcription of
the structural genes. The repressor molecule has been isolated and identified.
It is a so-called allosteric protein that can undergo changes in its three-dimensional
structure when it is combined with small molecules such as lactose. The
altered configuration of the repressor molecule accounts for the inability
of the repressor to bind to the operator gene.
Thirty-one operons have been identified in the E. coli linkage
map (11); therefore, the operon model is one of the
very important modes of regulation in bacteria. Some of these operons may
act in ways different from that of the lactose operon. However, all of
them involve the action of regulator and operator genes on the structural
genes. Whether the operon mechanism exists in eucaryotes is not known.
Understanding the gene expression control mechanisms is difficult because
the eucaryotic chromosome is a complex structure consisting of chromosomal
proteins and chromosomal RNA and DNA. Some data, though limited, suggest
that a type of operon exists (12). Mutation in regulator
genes in the eucaryotic chromosomes are thought by some to be the raw material
for molecular evolution. This concept will be considered further in the
following section.
[Section 2.6 Continued]



References 2.6
1. Griffith, F. J. Hyg. Camb. 27:113; 1928.
2. Avery, O. T.; MacLeod, C. M.; McCarty, M. J.
Exp. Med. 79:137; 1944.
3. Watson, J. D.; Crick, F. H. C. Cold Spring Harbor
Symp. Quant. Biol. 18:123; 1953.
4. Watson, J. D.; Crick, F. H. C. Nature. 171:464;
1953.
5. Nirenberg, M. W.; Mataei, J. H. Proc. Natl.
Acad. Sci. USA. 47:1588; 1961.
6. Nishimura, S.; Jones, D. S.; Khorana; H. G. J.
Mol. Biol. 13:302; 1965.
7. Watson, J. D. Molecular biology of the gene.
Menlo Park, CA: Benjamin; 1976: 374.
8. Lane, C. D.; Marbaix G.; Gurdon J. B. J. Mol.
Biol. 61:73; 1971.
9. Watson, J. D. Molecular biology of the gene.
482-83.
10. Jacob, F.; Monod, J. J. Mol. Biol. 2:318;
1961.
11. Bachmann, B. J.; Low, K. B.; Taylor,
A. L.
Bacteriol. Rev. 48:116; 1976.
12. Davidson, E. H.; Britten, R. J.
Q.
Rev. Biol. 48:565; 1973.















