Figure 2.53. Six distinguishable kinds of molecular mutation. (++), normal base pairs; (xx), substituted base pairs. See text for explanation.

2.6 Evidence from Molecular Biology and Genetics (continued)
2.6.3 Molecular Principles of Mutation. There are six distinguishable
kinds of molecular mutations caused by changes in the DNA (13)
(Figure 2.53).
Figure 2.53. Six distinguishable kinds of molecular mutation. (++), normal base pairs; (xx), substituted base pairs. See text for explanation.
1. Substitution of one or several nucleotidesa) Nucleotide Substitution. Even though there are six types of molecular mutations, it is likely that only nucleotide substitution is important in evolution. Most spontaneous mutations fall into this category. Many spontaneous mutations have little if any adverse effect on organisms, and sometimes they may even impart selective advantage.
2. Nucleotide deletion
3. Insertion of one or several nucleotides followed by restoration of sugar-phosphate bonds in DNA [159]
4. Removal of a segment of the polynucleotide chain (extended deletion)
5. Excision of a DNA segment and insertion of the segment at a different site (translocation)
6. Excision of a DNA segment and reinsertion of the segment at the same site of removal but with an 180° rotation (inversion)
Two ways that nucleotide substitutions come about is through transversion
and transition (Figure 2.54). Transversion is the replacement of
a pyrimidine with a purine or a purine with a pyrimidine. Transition is
the replacement of a pyrimidine with a pyrimidine or a purine with a purine.
Several mechanisms can account for these phenomena.
Figure 2.54. Classes of nucleotide replacement. When a mutation substitutes one nucleotide for another in a strand, at the next replication the new nucleotide is paired to its regular partner. The resulting pair is represented. Example: if in the upper TA pair, T is replaced by C, the result is the lower CG pair. Note that each base pair can undergo one kind of transition and two kinds of transversions. Reprinted, with permission, Davis, B. D. et al. Microbiology. Hagerstown, MD: Harper & Row, Publishers; 1973.
(1) Tautomeric Shift. Depending on the pH (-log[H+]) of the
medium, the bases in the DNA can exist as two or more forms by internal
rearrangement of hydrogen atoms. The process of internal rearrangement
is called tautomerization.
For example, thymine normally exists
in the keto form, allowing it to pair with adenine. Occasionally, tautomerization
produces the enol form of thymine allowing it to pair with guanine instead
(Figure 2.55). If tautomeric shift of thymine occurs during a round of
DNA replication, the resulting daughter strand of DNA will have acquired
a guanine in place of adenine. This is a transition mutation. [160]
Figure 2.55. Regular and unusual base pairing of thymine. I. Regular base pairing (in the common keto form) with adenine. II. Base pairing (in the rare enol form) with guanine. The heavy arrow in II indicates the displacement of the proton in the tautomerization of thymine. Reprinted, with permission, Davis, B. D. et al. Microbiology. Hagerstown, MD: Harper & Row, Publishers; 1973.
(2) Base Deamination. Many side products or intermediates
of cellular metabolism, such as peroxides, nitrous acid, formaldehyde,
and purine analogues, may be mutagenic and can cause transition mutation.
For example, nitrous acid can deaminate adenine, cytosine, and guanine
to hypoxanthine, uracil, and xanthine, respectively (Figure 2.56). Hypoxanthine
and uracil pair with different bases than adenine and cytosine. Deamination
of guanine does not affect the pairing specificity.
(3) Mutator Gene Effect. There is a mutation in the bacteria
E.
coli and Salmonella typhimurium that causes an increase in the
spontaneous mutation rate for all detectable genetic loci by a factor of
100 to 1000. A region of DNA called the mutator gene is responsible
for this event (14). [161] The mutations caused by the
mutator locus are all transversion mutations. The product of the bacterial
mutator gene has not been identified, but a similar mutator gene in the
bacterial virus coliphage T4 (15) has been known to
produce DNA polymerase, the enzyme responsible for the replication process
of DNA. Thus, a mutation in the mutator locus apparently alters the behavior
of DNA polymerase in such a way that the fidelity of DNA replication is
diminished, leading to transversions attributed to base mispairing. Although
there remains a possibility that mispairing can occur in the presence of
normal enzymes, it occurs at a very low rate (16).
The gene mu located in the third chromosome of the fruit fly Drosophila melanogaster may be the counterpart of the procaryotic mutator gene. It increases the rate of lethal mutations as well as mutations having morphological effects by suppressing mechanisms of chromosomal repair (17, 18).Figure 2.56. The oxidative deamination of DNA by nitrous acid, and its effect on subsequent base pairing. (a) Adenine is deaminated to hypoxanthine, which bonds tocytosine instead of to thymine. (b) Cytosine is deaminated to uracil, which bonds to adenine instead of to guanine. (c) Guanine is deaminated to xanthine, which continues to bond to cytosine. R is the sugar-phosphate backbone of DNA. [162]
(4) Irradiation Effect. Mutations caused by irradiation may occasionally be important in evolution. The best documented sources of irradiation are ultraviolet light and x-ray. Ultraviolet light is known to cause dimerization in adjacent pyrimidine bases in the DNA (Figure 2.57). The dimerized thymines will lose their pairing specificity. During subsequent DNA replication, gaps will be formed opposite to the thymine dimer in the DNA duplex that is not functional. Pairing up of the daughter duplexes may lead to recombination that will repair the gaps left by the dimers. But frequently, mistakes in base pairing may occur resulting in nucleotide substitution in the recombinant molecule (Figure 2.58). X-rays can also induce breaks in the DNA molecules as well as cross-linking within and between duplexes (Figure 2.59).
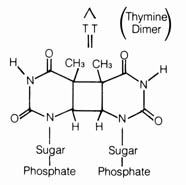
Figure 2.57. Thymine dimers produced by ultraviolet irradiation.
b) Nucleotide Deletion. Besides nucleotide substitutions, a few
spontaneous mutations have been identified as deletions of DNA segments.
The deletions may involve one or more genes. A good example is the tryptophan
dependent E. coli mutants that are also resistant to T1
coliphage (16). Genetic studies indicate that the DNA
of these mutants has a deletion of the segment containing the receptor
site for T1 coliphage and also the genes encoding the tryptophan
(an amino acid) biosynthetic [163] enzymes. The cause of the E. coli
deletion
mutants is not known.
2.6.4 Interrelationship of Molecular Mutations. It is often said that mutations are mostly harmful and are usually eliminated by selection; therefore, they cannot serve as the raw material for evolution. In most of the mutations involving drastic changes in the DNA, the mutants are lethal. Deletion, insertion, translocation, and inversion involving large segments of DNA will induce such massive alteration that all the triplet codes starting from the point of mutation will be changed, causing an almost complete revamping of the amino acid sequence of the gene product. The mutated protein will usually be nonfunctional, and the mutants will die.Figure 2.58. Ultraviolet-induced mutation. (A) Irradiation produces thymine dimers at random sites in the DNA molecule. (B) Replication produces a nonfunctional duplex with a gap opposite the dimer and a functional duplex from the strand with no thymine dimer. (C) Sister strand recombination reconstitutes an undamaged duplex. Base-pair changes or mutations (x x) occur as a result of errors in the recombination process. (++) are normal unmutated base pairs.
If the mutation changes only a small segment of the amino acid sequence that does not affect the function of a protein, the mutation is selectively neutral. For example, if the amino acid replaced due to nucleotide substitution has similar ionic properties as the original amino acid, the effect to the overall structure of the protein may be minimal, and the mutation will also be almost neutral.Figure 2.59. Possible mechanism by which cross-linking of the double helix of DNA can occur after x-ray irradiation. R1 and R2 are any organic molecules located on the opposite sides of the double helix. Thus a covalent linkage between R1 and R2 represents the crosslinking of the DNA molecule. [164]
Figure 2.60. The effect of two compensating frame-shift mutations in the gene coding for lysozyme in the bacteriophage T4. Only a fraction of the messenger RNA is shown. A nucleotide deletion has occurred in the third codon, and an addition in the seventh codon. As a consequence, the amino acid sequence is changed from the third to the seventh amino acid, but the normal sequence is restored after the seventh amino acid shown. X = A, G, C, or U; Y = A or G. Reprinted, with permission, from Dobzhansky, T. et al. Evolution. San Francisco: W. H. Freeman and Co.; 1977. © 1977 W. H. Freeman and Co.
Many potentially harmful mutations are suppressed by some interesting mechanisms. Translation mistakes can correct for mutation caused by nucleotide substitution by bringing in the right amino acid for the wrong codon, resulting in phenotypic suppression of the mutation. Finally, two mutations may restore the reading frame of the DNA and thus enable the gene product to become functional again if the segments changed by the two mutations only code for the nonessential part of the protein (Figure 2.60). Some examples that demonstrate the apparently beneficial effects of some mutations will be reviewed in the following section.
2.6.5 Nature of Chromosomal Mutation. Multiplication of eucaryotic
cells occurs by mitosis except for production of gametes that are produced
by meiosis (Figure 2.61; 2.62). During the early phases of mitosis and
meiosis, chromosomes are doubled (replicated) and then become visible as
the nuclear material condenses. Mitosis involves a single chromosomal separation
event to assure that each daughter cell receives the same genetic material
as the parent cell. In contrast, the events of meiosis [165] result in
genetic variability among the newly formed cells. Following the synapsis
of homologous chromosomes in meiosis, chromatid arms become entwined, leading
to recombination of genetic material through chance chromatid breakage
and reunion with the wrong partner (Figure 1.5). This process, called crossing
over, produces new genetic combinations in the daughter cells. [166]
The process of chromosomal replication and crossing over occasionally leads to chromosomal mutations. These mutations can be classified as structural chromosomal changes that affect the arrangement of genes in the chromosomes, and/or numerical changes that affect the number of chromosomes (19). The first category can be further subdivided into (1) changes due to loss or reduplication of some of the genes on the chromosome and (2) changes due to altered arrangements of the genes. Deletion and duplication involve chromosomal loss and gain, respectively. When the genes ABCDEFG carried by a normal chromosome have been cut to only ABEFG, a chromosomal deletion of the region containing genes CD has occurred. On the other hand, if the chromosome has acquired two additional genes and carries instead ABCDCDEFG, duplication has added the extra CD gene to the chromosome.
Figure 2.61. Mitosis. All cells go through these basic stages as they divide. Reproduced, with permission, from Moore, J. N.; Slusher, H. S. editors. Biology. Grand Rapids, MI: Zondervan Publishing House; 1970.
Figure 2.62. Meiosis. Formation of gametes (reproductive cells) is a process of chromosome reduction resulting in cells with one set of chromosomes. Reprinted, with permission, from Moore, J. N.; Slusher, H. S. editors. Biology. Grand Rapids, MI: Zondervan Publishing House; 1970.
Translocation, inversion, and transposition are patterns of altered arrangement of genes in the chromosome. Translocation involves the exchange of genes between two chromosomes. If chromosome 1 with genes ABCDEFG exchanges some of its genes with chromosome 2 with genes HIJK, a "new" set of chromosomes with genes ABCDJK and HIEFG may result. Inversion changes the location of several genes on the chromosome by rotating them 180° such that genes ABCDEFG will become AEDCBFG. Transposition simply moves the genes on the chromosome from one location to another as in the case of ABCDEFG to ADEFBCG.
There are several conditions that result from numerical changes in chromosome number. Aneuploidy results when the chromosome number is not an exact multiple of the single set of chromosomes in the sex cells. Polyploidy results from an even-numbered multiplication of chromosomes beyond that of the diploid configuration of most cells.
Chromosomal mutations are observed in natural populations, but they are usually harmful to the organisms. Although Goldschmidt proposed that systematic chromosomal rearrangement (20) can give rise to a "hopeful monster" under certain environments, his thesis has not been confirmed by experimental observations.
Polyploidy is commonly found in plants and constitutes rare examples of speciation that can be directly observed (see I.3.3.1.c). However, its significance in the evolution of animals is doubtful because the majority of the animals that have been reported to be polyploids are parthenogenetic, i.e., they developed from unfertilized eggs. Thus, polyploidy fails to account for the evolution of bisexualism and the genetic diversity in animals, for it cannot be established in species with separate sexes and regular outcrossing. [167]
Translocation can give rise to chromosomal duplication and deletion
during meiosis (Figure 2.63). In plants, pollen grains and ovules with
duplicated or deleted chromosomes are usually nonfunctional, though there
is evidence that translocation is regularly tolerated in some species.
Figure 2.63. Chromosome duplication and deletion arisen from translocation during meiosis. Genes a, b, c, d, and e, f are genes on two separate pairs of homologous chromosomes.
[168] Animal gametes with duplicated or deleted chromosomes may
function, but the zygote formed by the union of a normal gamete and one
of these defective gametes will usually die or develop into an abnormal
individual. Therefore the possibility that chromosomal mutations can provide
the raw material for evolution is very much in doubt.
2.6.6 Evolution and Genetic Equilibrium. The Mendelian concept of dominant and recessive genes raises an interesting question when dealing with a population of interbreeding individuals with different genotypes. Will the recessive genes be eventually replaced by the dominant genes? The search for the answer to this question led G. H. Hardy, an English mathematician, and G. Weinberg, a German physician, to develop the Hardy-Weinberg model of genetic equilibrium. They defined the relative proportions of genes and genotypes in a population as the gene frequency and the genotype frequency, respectively. The principle of genetic equilibrium is that gene frequencies and genotype frequencies will stay constant in a population under the following conditions:
Equation (2) is actually the binomial expression of (p + q)2, and thus, it is an expansion of equation (1).p + q = 1 (1)
p2 + 2pq + q2 = 1 (2)
Let us consider an example of a hypothetical population that is composed
entirely of heterozygous individuals (Aa). Assuming random mating,
the proportion of the genotypes in the F1 offspring will be ![]() AA
:
AA
: ![]() Aa
:
Aa
: ![]() aa, (see I.1.3).
The gene frequencies of A and a in the parental generations
are both 0.5. However, the gene frequencies in the [169] F1
generation are less obvious. To calculate the gene frequency of A (p)in
the F1 generation proceed as follows:
aa, (see I.1.3).
The gene frequencies of A and a in the parental generations
are both 0.5. However, the gene frequencies in the [169] F1
generation are less obvious. To calculate the gene frequency of A (p)in
the F1 generation proceed as follows:

Substituting q and a in the above equation gives the frequency of the a allele.
In the second generation, random mating will give rise to the results listed in Table 2.17 with the genotype distribution of 4/16 AA : 8/16 Aa : 4/16 aa. The gene frequencies of A and a can be again calculated by using equation (3) to be 0.5 and 0.5.
Thus, we have demonstrated that the gene frequencies of A and
a
remain
constant in the parental, F1 and F2 generations.
The genotype frequencies of AA, Aa, and aa also remain the
same in the first and second generation as predicted by equation (2):
p2 = AA = (0.5 x 0.5) = 0.25Table 2.17. The Offspring of the Random Mating of a Population Composed of 1/4 AA, 1/2 Aa, and 1/4 aa Individuals.*
2pq = Aa = (2 x 0.5 x 0.5) = 0.50
q2 = aa = (0.5 x 0.5) = 0.25*NOTE: Reprinted, with permission, from Villee, C. A. Biology. 7th ed. Philadelphia: W. B. Saunders Co.; 1977. © 1977 by the W. B. Saunders Co.
[170] The gene frequencies will remain constant for an infinite
number of generations as long as the four basic conditions listed earlier
are maintained.
Therefore, Hardy and Weinberg predicted that there will be no change in gene frequencies and genotype frequencies regardless of the dominant or recessive characters of the genes as long as the four basic conditions are met. The principle of genetic equilibrium has provided a useful tool to monitor the most fundamental step of evolution — the change in gene frequencies (microevolution). Since there are very few natural populations that have maintained all of the four conditions of the Hardy-Weinberg law, it may be predicted that evolution is constantly occurring.
Several factors may contribute to the change of gene frequencies, among them are mutation pressure, selection pressure, and genetic drift.
a) Mutation Pressure. Mutation pressure is the difference in rate of forward mutation verses backward mutation. Since spontaneous mutations are always occurring and mutation equilibrium is seldom achieved in the natural population, mutation pressure tends to cause a slow "shift" in the gene frequencies in the gene pool favoring the more stable alleles over the more mutable alleles.
In spite of the popular belief that mutation is the raw material of evolution, since the mutation rate is so slow (on the order of 10-6 per generation), it alone seldom exerts much influence on evolution with the following exceptions: (1) microorganisms that have short generation times, (2) some higher plant groups in which polyploidy contributes to rapid speciation, and (3) small populations subject to genetic drift. In all the above cases, mutations are random and appear to influence only slightly the nature and direction of evolution.
b) Selection Pressure. Selection pressure is by far the most
important factor directing the change of gene frequencies in a population.
It can change the frequency of a particular gene drastically by selecting
for or against it in every generation. Consider a hypothetical population
in which the initial frequencies of alleles A and a are 0.9
and 0.1 and the genotype frequencies AA, Aa, aa are 0.81, 0.18,
and 0.01, respectively. If A is selected against by a selection
pressure such that its frequency is reduced to 0.8 in the same generation,
then the frequency of a becomes 0.2 and the genotype frequencies
of AA, Aa, and aa will be changed correspondingly to 0.64,
0.32, and 0.04 (Table 2.18). If the same selection pressure against A
is maintained throughout subsequent generations, the frequencies of A
and AA will be reduced drastically to a barely detectable level.
Such selection will lead to the wholesale revision of the frequencies of
the genes and genotypes in the population. It can change the genetic [171]
structure of the population in the absence of mutation.
Table 2.18. Comparison of genotype frequencies under selection and no selection. See text for explanation.
However, if new competing alleles are produced through mutation,
they may be selected for by a changing environment. Eventually the new
genes and genotypes may be established by natural selection. Examples of
evolutionary change brought about by selection are found both in laboratory
studies and in natural populations (DDT resistance, antibiotic resistance,
sickle cell anemia). They will be discussed in 3.2.1.b, c. The three types
of selection — stabilizing, directional, and disruptive (see I.1.4)
— interact with each other constantly and probably account for most of
the microevolutionary changes observed in nature.
c) Genetic Drift. Sudden random change of gene frequencies can occur in a population of any size, but it will produce the greatest fluctuations in small populations characterized by little or no migration.
The effect of genetic drift can be illustrated by a simple example. We can compare the ability of individuals in two hypothetical populations to taste the chemical phenylthiocarbamide (PTC). One population consists of 100 persons and the other 10,000. If the frequencies of the dominant nontaster allele T and the recessive taster allele t are 0.6 and 0.4, respectively, in the parental generation of both populations, then the genotypic frequencies of TT, Tt, and tt will be 0.36, 0.48, and 0.16, respectively. This means that 16 individuals in the small population and 1600 individuals in the large population will have the homozygous recessive genotype tt, the taster phenotype. If by accident eight individuals from each population die in a car crash, the gene frequency of the t allele will be [172] changed significantly in the small population but not in the large population. This can be seen by tabulating the gene frequencies as follows:
For the small population, the genotype distribution after the trauma will be:
The gene frequency of t (q) after the trauma is calculated from equation (3) as:
The corresponding frequency of T (p) is
p = 1 - q = 1.0 - 0.348 = 0.652By the same token, the gene frequencies of t and T in the large population after the trauma will be 0.3995 and 0.6005. Thus, only the small population experiences a significant change in gene frequencies caused by the chance event.
Genetic drift can also be caused by inbreeding in a small population that can be represented by the founder effect. If a population is founded by a few individuals being isolated from the rest of the outbreeding population, it is possible that the isolated individuals may have unusually high frequencies of one or more harmful alleles. These alleles presumably result from previous inbreeding. Founder effect may plague also the surviving members of a population that suffers a famine or other catastrophes. The expansion of such a small population could lead to the establishment of harmful genes, the so-called bottle neck effect.
Examples of genetic drift can be found in laboratory and natural populations (21). It has been shown that the variability of chromosome arrangement of experimental populations of Drosophila pseudoobscura raised in isolated conditions depends on the number of founding individuals carrying different chromosome gene arrangements. The genetic homogeneity of certain human populations can also be traced to the few founders. For example, the Ramah Navaho Indian tribe began their population after isolation from an outbreeding population.
Finally, one must consider the interplay of neutral mutations and genetic drift resulting in gene frequency change. Mutations that are neither beneficial nor harmful to organisms are termed neutral and are the source of new genes that will be established eventually in the population by [173] random drift. In other words, there is no selection exerted on the mutations by the environment or from intrinsic factors within the population.
Recently it has been hypothesized that a large amount of genetic variability may be attributed to genetic drift acting on neutral alleles formerly produced by a substantial number of neutral mutations. Aspects of this hypothesis will be explored in I.3.3.2.a.1.
References 2.6