


3.3 Evaluation of Macroevolution
Despite the systematic unity and comprehensive scope of macroevolution
(synthetic or general theory of evolution), it has serious weaknesses when
evaluated.
3.3.1 Empirical Inadequacy.
a) Demise of the Theory of Spontaneous Generation. The theory
of spontaneous generation states that life arises continually from the
nonliving, and this idea was accepted by most medieval scholars as a mechanistic
explanation of the origin of life. Aristotle, Newton, William Harvey, Descartes,
and van Helmont are but a few of a long list of learned men who accepted
certain forms of the theory of spontaneous generation. Some scientists
with a more naturalistic outlook of life chose to accept the theory as
an alternative to the belief in a single primary act of a supernatural
Creator.
The theory of spontaneous generation had its formal beginning after
Anton von Leeuwenhoek (1632-1723) revealed to the scientific world the
vast number of microscopic organisms in nature. The ancient thought that
many plants and animals can be generated spontaneously under special conditions
immediately took up a new dimension and presented itself in the form of
the abiogenesis of microscopic organisms. This theory was the predominant
view among scientists at that time because of its apparent consistency
with the interpretation of observations that many organisms sprang from
apparently nonliving material of various kinds.
There were always skeptics who questioned the popular view and ventured
to test the theory of spontaneous generation experimentally. Francesco
Redi tested the theory that maggots were derived from putrid meat by spontaneous
transformation of some of the meat's components (see I.3.1). Redi
proved this hypothesis untenable by demonstrating that maggots appeared
to arise from meat only if flies had laid eggs on the meat. Redi's results
and other studies of the origins of plants and animals weakened the theory
of spontaneous generation considerably. However, since microorganisms were
difficult to handle because of the primitive [192] microbiological techniques
used at the time, the proponents of the theory focused their attention
on the origins of these microscopic "beasts" in organic infusions.
Louis Joblot paved the way for the eventually successful refutation
of the theory of spontaneous generation by demonstrating that a heated
hay infusion in a closed vessel did not give rise to microorganisms, but
an unheated infusion in an open vessel was teeming with them. Lazzaro Spallanzani
later showed that the same phenomena could be observed in heated meat broth.
However, his results differed from those of another scientist John Needham,
who had earlier found that life developed in broth in a heated closed vessel
as well as in an open unheated vessel.
Needham, a proponent of the abiogenesis of life, criticized Spallanzani's
experiment by alluding to the effect of prolonged heating that might have
destroyed the "vital force" in the broth that was necessary for the abiogenesis
of microorganisms. Spallanzani answered Needham's challenge by showing
that the heated broth could support the growth of microorganisms after
it was reexposed to air. However, the supporters of abiogenesis persisted
in their criticisms by arguing that it was the exclusion of oxygen, which
they believed was essential for the growth of the microorganisms, from
the air in the closed vessel that prevented the abiogenesis of life in
Spallanzani's experiments. They concluded that the appearance of microorganisms
in the heated broth after it was reexposed to air was simply the result
of the reintroduction of oxygen into the vessel.
Although Spallanzani's interpretation of his experimental results was
not accepted by many of his contemporary scientists, the finding enabled
Francois Appert to develop a canning method to preserve perishable organic
materials. Appert enclosed food in airtight cans, heated the containers,
and found the food could be preserved indefinitely. At the same time, the
argument of the exclusion of oxygen continued to be used by the proponents
of the theory of spontaneous generation to explain away Appert's results.
Furthermore, proponents continued to amass erroneous data from poorly controlled
experiments to support their contention.
It was not until 1861, when Louis Pasteur (1822-95) presented unequivocal
empirical evidence, that the theory of spontaneous generation was finally
discredited. Pasteur first demonstrated that air does contain microorganisms.
He filtered air through a tube plugged with a piece of guncotton. The guncotton
was then dissolved in a mixture of ether and alcohol. Microscopic examination
of the sediment of the guncotton solution revealed the presence of microorganisms.
Pasteur showed also that the introduction of heated air into a boiled sterile
infusion in a closed system did not cause microbial contamination. However,
if germ-laden [193] guncotton obtained by filtering air was added to it,
the infusion soon teemed with microorganisms.
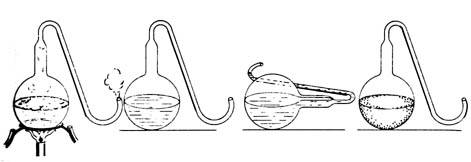 Figure 3.8. Pasteur demonstrated that bacteria were trapped
in the curved neck of the flask by showing that as long as the flask with
the boiled broth remained upright no decay occurred. But when the flask
was tipped causing the broth to enter the neck and then set upright, the
broth quickly showed signs of bacterial growth. Reprinted, with permission
from Moore, J. N.; Slusher, H. S. editors. Biology. Grand Rapids:
Zondervan Publishing House; 1970.
Figure 3.8. Pasteur demonstrated that bacteria were trapped
in the curved neck of the flask by showing that as long as the flask with
the boiled broth remained upright no decay occurred. But when the flask
was tipped causing the broth to enter the neck and then set upright, the
broth quickly showed signs of bacterial growth. Reprinted, with permission
from Moore, J. N.; Slusher, H. S. editors. Biology. Grand Rapids:
Zondervan Publishing House; 1970.
Pasteur designed a specialized set of growth chambers, the swan-necked
flasks, to show that filtration through cotton was not necessary to
eliminate microbial contamination (Figure 3.8). The long curved neck prevented
dust particles carrying microorganisms from entering a flask. Although
air exchange between the broth chamber and the outside atmosphere could
occur through the neck, heated broth was free from microbial contamination
because microorganisms from the air were trapped on the inside walls of
the necks by gravity. Interestingly, the broth could be contaminated by
tilting the flask allowing the broth to wash the inside of the neck and
then drain back into the flask or by breaking off the neck near the body
of the flask, allowing contaminated air to enter.
John Tyndall gave the final death blow to the classic version of the
theory of spontaneous generation. Using a box similar to that depicted
in Figure 3.9, Tyndall demonstrated that dust-free air would not contaminate
sterile broth. The entire interior of Tyndall's box was coated with glycerine
to trap microorganisms. The box was practically sealed off from the atmosphere
except for the two openings on either side of the box connected to convoluted
tubings that allowed air to enter but trapped dust. After standing several
days, the dust particles, floating in the air inside the box settled on
the bottom, and air became "optically empty" (or revealed no suspended
articles in the beam of light shone through the window) (see Figure
3.9). Then a pipette was inserted through a rubber stopper on the top of
the box, and meat broth was introduced through the [194] pipette into the
tubes. The tubes were then boiled by immersing them for long periods in
boiling brine. Tyndall found that after broth in the tubes had been cooled
down to room temperature, it could be kept free of microbial growth indefinitely.
Thus, his experiment eliminated the theory of spontaneous generation.
However, since the mechanistic outlook of life has never recognized
any essential qualitative difference between the inorganic world and living
organisms, the direct transformation of the inorganic world to the living
world through some sort of spontaneous generation is necessary for this
point of view. This apparently motivates the ardent supporters of the theory
of spontaneous generation to fight to the very end.
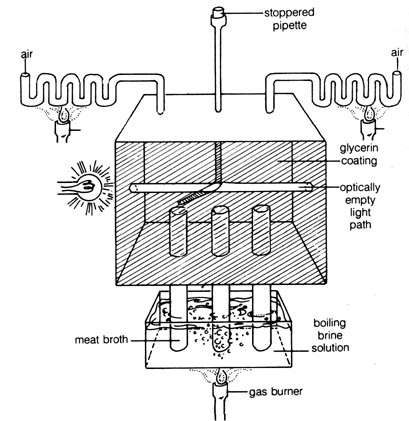 Figure 3.9. A diagram of the box that Tyndall used to demonstrate
that "optically empty" air contained no microorganisms.
Figure 3.9. A diagram of the box that Tyndall used to demonstrate
that "optically empty" air contained no microorganisms.
Against the completely insurmountable wall of facts, some proponents
of the mechanistic view gave up their ideas of spontaneous generation and
[195] instead claimed that life emerged on earth by the transport of seeds
from another world (panspermia). Proponents of panspermia believe
that life forms were brought to earth by meteorites and/or cosmic dust.
Numerous
attempts to isolate microbes from meteorites have failed. Pasteur also
tested this hypothesis by trying to isolate viable bacteria from a carbonaceous
meteorite, but he was unsuccessful (1). Alleged success
in isolating microbes from meteorites cannot be verified. Improper experimental
techniques were used, including failure to avoid outside microbial contamination.
Transfer of living spores by cosmic dust from one heavenly body to another
under the pressure of stellar rays is also an untenable theory to account
for the origin of life on earth. The energetic irradiation penetrating
interplanetary and interstellar space is destructive to all life, and it
is inconceivable to maintain the viability of living spores or seeds on
cosmic dust particles.
Finally, there is the speculation that life might have been brought
to earth some time ago by interplanetary or interstellar travelers similar
to our cosmonauts and astronauts. This remains at the level of science
fiction, for it has no factual basis.
The majority of scholars who hold to a mechanistic origin of life have
turned their attentions to a modified version of the theory of spontaneous
generation. Their reasoning is as follows: Since life existed during only
part of the earth's history, and since a divine act of creation is untestable
and thus unscientific, life must have originated early in the earth's history
by spontaneous generation under a different set of conditions, but this
no longer happens. The idea of spontaneous generation under a different
physical environment provides a way out for the mechanists. They can simulate
the presumed primordial conditions of the earth in the laboratory
and then test the possibility of the abiogenesis of life.
The mechanists believe that the last missing element of the general
theory of evolution linking the first cell to inorganic matter has been
supplied by the currently popular theory of spontaneous generation. However,
the stipulation of the abiogenesis of life under conditions different
from
the present has removed the theory from the realm of empirical sciences,
for it can neither be verified nor falsified by experiments done under
present earth conditions (2). In addition, there are
difficulties in the experimental documentation of the spontaneous generation
of the first cell. These problems will be discussed in the following section.
b) Difficulties in Accounting for the Abiogenesis of the First
Cell. The most commonly held theories of abiogenesis of life assume
three [196] stages of chemical evolution (Figure 3.10). The first stage
involves the accumulation of certain organic molecules due to the random
processes of collision and irradiation in the primeval "soup" of the ocean.
The second stage is the selection of certain thermodynamically stable
organic molecules existing in clusters or the formation of macromolecules,
such as proteins and nucleic acids. In the final stage of chemical evolution
some of the macromolecules acquire the capacity to reproduce (replicate),
using some type of template mechanism that is the beginning of the
first cell. Mechanists believe all of these chemical reactions were carried
out in a reducing atmosphere devoid of oxygen since an oxidizing atmosphere
would quickly destroy large molecules that formed.
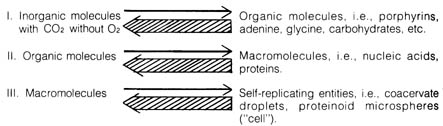
Figure 3.10. THREE STAGES OF CHEMICAL EVOLUTION.
The above stages are thought possible for the following reasons:
(1) The earth and solar system are thought to be the results of the condensation
of a cloud of cosmic dust rich in hydrogen; therefore, the primeval atmosphere
of the earth must have contained this gas. Furthermore, the reducing atmosphere
of hydrogen, methane, and ammonia observed on several planets of the solar
system such as Jupiter, Saturn, Uranus, and Neptune supports this assumption.
(2) It is assumed that the composition of meteorites is similar to that
of the primeval earth. All the elements analyzed exist in meteorites in
reduced form; therefore, the primitive earth may also have been devoid
of the oxidizing agent oxygen. (3) Under the primordial conditions, many
experiments designed to synthesize bio-organic molecules failed when molecular
oxygen was present; however, they succeeded when oxygen was removed. Due
to these stipulations, it has been suggested that the first living cell
resulting from chemical evolution was an anaerobic cell existing deep in
the ocean, removed from ultraviolet solar irradiation that would destroy
it. However, Donald England (3) has made potent criticisms
on the experiments supporting each of these stages of chemical evolution.
Furthermore, an alternative model for the primitive atmosphere with ingredients
similar to today's [197] atmosphere but without oxygen has been proposed
(4).
This model also necessitates the stipulation of an anaerobic cell.
The most famous example of abiogenesis of organic compounds in stage
one is the experiment performed by S. L. Miller. He synthesized amino acids
by passing an electric discharge for seven days through a closed system
(Figure 3.11) containing methane, ammonia water, and hydrogen. Porphyrins,
important structural components of the photosynthetic and respiratory apparatus
of living cells, were also obtained in a similar manner. Adenine, an important
base in nucleic acids, was formed by chemical polymerization of hydrogen
cyanide and ammonia. Carbohydrates, including the sugar backbones of nucleic
acids, were also synthesized by incubating formaldehyde with an inert polar
polymer, alumina, in the presence of some naturally occurring minerals.
Miller's results show that carefully controlled experiments in a closed
system do result in the synthesis of a large variety of bio-organic compounds
identical to those found in the living cell. However, compounds were synthesized
only when sufficient starting materials were incubated with the right kind
and right amount of energy in a closed system. On the other hand,
in the primordial earth's open system with all processes random,
the synthesis of these bio-organic compounds by chance alone is
extremely improbable.
The second stage of chemical evolution involving the spontaneous origin
of macromolecules seems contrary to the second law of thermodynamics. The
law states that structures within a closed system tend toward a state of
maximum disorder or randomness (increase in entropy). Thus a structure
cannot become more complex (decrease in entropy) without the concomitant
dismantling of another structure (increase in entropy) such that the resulting
entropy of the whole system (i.e., the complex structure plus the dismantled
structure) is increasing.
A living cell can order its amino acids and nucleic acid bases into
proteins or nucleic acids, respectively. It does so by the efficient mechanisms
of gene expression and replication fueled by a large expenditure of energy
that has to be supplied by the breakdown of complex molecules acquired
as nutrients from the environment. Carbohydrates represent one such energy
source. During this process, the entropy of the universe increases slightly;
therefore, the cell does not violate the second law of thermodynamics.
During the earth's primordial condition, such an intricate energy conversion
machine as the cell did not exist. In isolated cases, energy may have been
expended from the surrounding to fuel the ordering of certain organic molecules
into bio-macromolecules. Yet the overall tendency in [198]
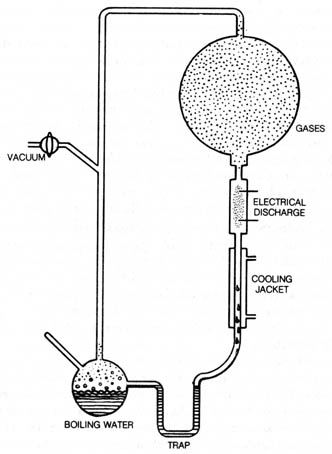
Figure 3.11. Experiment of S. L. Miller made amino acids
by circulating methane (CH4), ammonia (NH3), water
vapor (H20), and hydrogen (H2) past an electrical
discharge. The amino acids collected at the bottom of apparatus and were
detected by paper chromatography. Reprinted, with permission, from Wald,
G. The origin of life. Sc. Am. August 1954.
the primeval condition must have been such that spontaneous dissolution
of
transient macromolecules was much more probable than spontaneous
sustained
synthesis.
These stipulations are represented in Figure 3.10 by the heavy backward
arrows as compared to the slender forward arrows. In other words, the conditions
that are presumably necessary for the synthesis of bio-organic and macromolecules
are even more effective in decomposing them. Ultraviolet irradiation
that was assumed to be a primary energy source for abiogenesis cleaves
bonds of carbon compounds and causes their decomposition. Therefore the
amino acids formed by processes [199] analogous to Miller's experiment
would be subject to the forces of dissolution right after their synthesis.
The reason life can exist presently with these harmful solar irradiations
is that ozone in the upper atmosphere filters out most ultraviolet radiation
from sunlight before it reaches the earth. However, the ozone is formed
from molecular oxygen that was presumably absent in the primeval atmosphere.
The lack of protection of the primitive earth's surface from solar irradiation
by atmospheric gases prompted mechanistic evolutionists to suggest that
life-requiring organic compounds were synthesized in the stratosphere where
gases were diffused. This would allow organic compounds to be scattered
right after their synthesis and minimize the dissolution effects of ultraviolet
light.
It has been calculated that the rate of decomposition of glycine, the
most abundant amino acid synthesized in Miller's experiment, is much greater
than the rate of its formation, assuming the primordial conditions of the
earth as proposed by Miller and his collaborator Urey. Thus 97% of the
glycine synthesized in the atmosphere would be decomposed before it could
reach the earth's surface (5). The minute quantity of
glycine reaching the earth then must diffuse to a depth of at least 30
feet beneath the ocean's surface in order to escape the potential decomposing
effects of ultraviolet light. Therefore, it is easily seen that the amount
of organic compounds that could finally accumulate in the ocean would be
much less than what the evolutionists would expect in the "rich primordial
soup of organic nutrients," if they were accumulated at all.
The instability of covalent linkages in proteins, nucleic acids, and
carbohydrates also adds to the seemingly insurmountable barriers that have
to be overcome by advocates of abiogenesis. A. L. Lehninger stated: "In
order for primordial polypeptides, polysaccharides and polynucleotides
to accumulate in any amount in the primordial sea or in localized aqueous
systems, the rate of their formation must have exceeded the rate of their
degradation" (6). However, this stipulation is contrary
to what is expected from the second law of thermodynamics. Therefore, a
serious paradox exists in stage two of chemical evolution and cannot be
easily ignored.
The commonly cited examples of self-replicating and metabolizing prebiotic
systems (7) consist of coacervate droplets and
proteinoid
microspheres.
Coacervate droplets are made up of bio-organic compounds
of cell size in which organic macromolecules (amino acids, sugar, and bases)
are entrapped in polymeric forms in aqueous (watery) droplets. The tendency
to undergo coacervation is primarily a function of the molecular size and
the matrix structure of the polymer that allows the penetration of water
molecules. The droplet may increase in size to its physical limit and [200]
then break into two smaller droplets just as oil breaks into smaller droplets
when it is shaken in an aqueous environment.
Coacervate droplets are thought to be able to entrap a catalyst as well
as a substrate and thus become a site for a primitive one-reaction metabolism.
The droplets have been made in the laboratory from gelatin gum arabic,
ribonucleic acid, nuclear protein, and serum albumin. However, coacervate
droplets are unstable and lack the rigid template mechanisms that are typical
of the genetic material of a living cell. Therefore, it is far from an
adequate model of the first cell.
Proteinoid microspheres are synthesized when a high concentration of
aspartic and glutamic acids in a nearly anhydrous (waterless) condition
are heated to 170°C. Peptide bonds are formed, and proteinlike compounds
with molecular weights of 3000 to 11,000 are generated. When these compounds
are cooled slowly over a period of one or two weeks at the right pH and
salt concentration, spherical droplets about 2.0  (
( = micron) in diameter appear.
If the pH is adjusted properly, the outer boundaries of these microspheres
show double-layered structures resembling a cell membrane, However, the
outer boundary contains no lipid, an organic molecule always found in cell
membranes. On the other hand, the microspheres have been observed to undergo
"budding" or "cleavage" (processes common to living cells) when they are
allowed to stand for a long period, or if exposed to Mg++, or
if there is a shift in pH.
= micron) in diameter appear.
If the pH is adjusted properly, the outer boundaries of these microspheres
show double-layered structures resembling a cell membrane, However, the
outer boundary contains no lipid, an organic molecule always found in cell
membranes. On the other hand, the microspheres have been observed to undergo
"budding" or "cleavage" (processes common to living cells) when they are
allowed to stand for a long period, or if exposed to Mg++, or
if there is a shift in pH.
The major deficiency of proteinoid microsphere formation as a model
of the first cell is the absence of genetic material in the form of nucleic
acid capable of self-reproduction through replication. Proteinoid microspheres
would somehow have to be led to the formation of primitive nucleic acids
for the propagation of the first cell if the model is valid (8).
This process is difficult to accept since no present-day counterpart has
been observed.
Dr. Peter T. Mora, a leading authority in research on the origin of
life, has made some salient observations on problems faced by advocates
of the theory of chemical evolution (9). His remarks
are summarized in four points. The first point is that polymerization of
chemical monomers under simulated primordial conditions contains no more
than "information" input defined by physical and chemical means such as
in experiments of organic chemistry, and it does not start new life processes
capable of self-reproduction. Therefore, the results are analogous to the
self-assembling process of a computer that operates only to the extent
of information it is given. Mora's second point states that it is difficult
to account for the switch to a self-reproducing internal control
characteristic of the cell when chemical polymerization in the chemical
evolution models is thought to be triggered by external forces.
[201]
In Mora's third point he deals with selectivity. Used in the physiochemical
sense, selectivity does not parallel Darwinian selection that can explain
only how a "living system" with a capacity to adjust to its changing
environment reproduces persistently.
Selection, as used by Horowitz, Oparin, and other proponents of the
theory of chemical evolution, includes the assumption that the more probable,
less complex chemical events led to the acquisition of the more complex,
less probable events having increased stability. In the physiochemical
sense, selectivity can only mean the in vitro chemical reaction
that operates only when the "selective" conditions exist, i.e., the reactants
are in the right energy state, reactants collide, and catalysts are available.
None of the above conditions persist and thus cannot lead to a consistently
self-maintaining and self-reproducing system. The conditions produce only
a temporary metastable order or function that will cease and tend to disperse
more and more as its complexity increases. Therefore, natural selection
in the Darwinian sense cannot be applied at the molecular level.
Mora's last point states that living systems require an increase in
complexity and interaction of molecular aggregates. However, the presence
of random physiochemical forces operate to decrease the formation and interaction
of the above complexes. Therefore, there is a low probability that interacting
chemical systems will reproduce persistently and overcome disruptive changes.
The logical conclusion suggests that the origin and continuance of life
on earth is not controlled by the above principles.
c) Weak Empirical Documentation of Evolution Above the Species Level
(Macroevolution, Transpecific Evolution). Macroevolution (transpecific
evolution) above the species level rests quite heavily on the concept of
speciation. Although a rational explanation can be formulated to account
for diversification of species in nature by microevolution (see I.3.2.2),
it has not been observed in a controlled laboratory setting.
Experiments with the chemostat (see I.3.2.1) can allow the observation
of numerous generations of bacterial evolution in a relatively short period
of time. However, only varieties within a species do interchange genetic
materials, and no new species have been detected. Ernst Mayr (10)
pointing out the difficulty stated, “Knowing that there are alternative
modes of speciation, the student of evolution is faced by a methodological
difficulty. Speciation is a slow historical process and except in the case
of polyploidy, it can never be observed directly by an individual observer."
Polyploidy is a major phenomenon in plant evolution, and it can be observed
empirically. This phenomenon of plant speciation will be considered more
carefully (11) in the following discussion. [202]
Polyploidy was discovered 60 years ago when the chromosome numbers of
some plants were analyzed. The diploid numbers of plant chromosomes range
from 4 to well over 200. However, the most frequent number was 12, while
8 was the next frequent. About 50% of all plants have chromosome numbers
below 12. Plants with higher chromosome numbers usually have multiples
of the lower ones. Within a single genus, there is usually a series of
species in which the chromosome numbers of some are multiples of that of
another species. This condition in which the number of chromosome sets
in the nucleus is a multiple (greater than two) of the haploid numbers
is called polyploidy. This could happen in two ways, either a single set
of haploid chromosomes is present more than twice (autopolyploidy) or two
or more sets of chromosomes from different species are present, making
a total of more than two genomes (allopoyploidy). Allopolyploidy is more
frequent in plant speciation.
Autopolyploids are known both in nature and in experimental materials.
One of the bases of de Vries's mutation theory was an autopolyploid mutant
strain of Oenothera lamarckiana (evening primrose). It is a tetraploid
containing 28 chromosomes instead of the diploid number of 14, and other
tetraploid plants species exhibit features similar to those of this plant.
First, it is considerably larger than the diploid O. lamarckiana, and
de Vries named this autopolyploid plant Oenothera gigas because
of its size and regarded it as a new species. Second, the stems are thicker,
and the leaves are shorter, broader, and thicker than those of the diploid
plant. Third, it seems to have a slower growth rate than the diploid O.lamarckiana.
However,
this is not typical, because most tetraploids can adapt to more severe
environments. This is due to tetraploids usually being more vigorous than
their diploid counterparts.
An autopolyploid produces offspring that can be mated to the diploid
parent. Therefore an autopolyploid is not considered a new species. But
there is considerable reproductive isolation between a diploid and its
autotetraploid because the hybrid between them is a triploid (three haploid
genomes in each cell). Triploids are highly sterile because they
usually do not form regular gametes during meiosis. Many autopolyploids
with odd numbers of the haploid genome are nonviable. Therefore
the role played by triploids derived from autopolyploidy in plant speciation
is minimal.
Allopolyploidy also has been produced experimentally and observed in
nature. Two mechanisms are advanced to account for the occurrence of allopolyploidy
in plants and are based on the occasional failure of reduction division
during meiosis observed in plants and especially frequent in plants with
chromosome complements that do not synapse readily. [203]
In examining allopolyploidy, A and B will each represent
a different haploid chromosomal set. Thus the chromosomal segregation and
assortment in the cross of AA x BB will result in a hybrid
F1 of AB. In subsequent generations, if there is insufficient
homology between A and B to permit synapsis, a significant
percentage of AB gametes may be produced in meiosis, due to the
failure in reduction of chromosomes. In a self-fertilized plant, some AB
ovules
will be fertilized by AB pollens. Thus an allotetraploid
AABB
is
formed at once. This allotetraploid has two sets of homologous chromosomes,
and there is little tendency for synapsis to occur between
A and
B
(a
condition that leads to this formation in the allotetraploid in the first
place). Also, there is little chance to have a complex and irregular segregation
pattern during meiosis. Therefore the allotetraploid is perfectly fertile.
There is another mechanism by which allopolyploids can be formed. This
mechanism involves a two-step utilization of the failure of reduction during
meiosis. If a hybrid AB is backcrossed with one of its parent AA,
an
occasional nonreduction may occur, and AB fails to segregate during
meiosis. Thus the offspring becomes AAB. If AAB is then mated
with BB and fails to segregate during meiosis, then the allotetraploid
AABB
is formed. Allotetraploid may also be formed by accidental doubling
of the chromosomes in the zygote of the original hybrid AB,
in a
manner analogous to the experimental induction of chromosomal doubling
by treatment with the drug colchicine. This drug blocks the assembly of
the mitotic spindle apparatus.
Allotetraploids show characteristics of both parental species, in addition
to new tetraploid characteristics. The species are good because they can
propagate indefinitely without any apparent defect in their reproductive
machinery. They are also reproductively isolated from their parent by the
sterility or inviability of the hybrid produced by the cross between the
allotetraploid and either of the parent species. This can be visualized
by examining the meiotic pattern of the hybrid AAB obtained from
mating AABB and AA. The two A chromosome sets can
form a synaptic pair and can undergo reduction in meiosis. However, the
B
set
does not synapse, and it will be distributed randomly to each gamete. Thus
it has meiotic difficulties and is highly sterile.
Allopolyploidy is commonly observed in nature and is one of the major
mechanisms of plant speciation. It is a good example of sympatric speciation
(see I.1.5). The evolution of bread wheat is a classic example of
sympatric speciation through allopolyploidy. Modern wheat (Triticum aestivum)
is a hexaploid represented by AABBDD (Figure 3.12). Its lineage
can be traced to the tetraploid wheat Triticum dicoccum with an
[204] AABB genome that is produced by the intergeneric cross between
the diploid wheat Triticum monococcum (AA) and goat grass Aegilops
speltoides (BB). Later, a second intergeneric cross between T. dicoccum
and Aegilops squarrosa, the latter contributing the D genome,
occurred to produce modern bread wheat.
Although polyploidy plays a major role in plant speciation, it is considered
to be of minor significance in animals. The majority of the animals that
have been reported to be polyploids are parthenogenetic (reproduction without
fertilization) or hermaphroditic (having both sexes in the same individual).
All of the parthenogenetic organisms are arthropods such as the water flea,
brine shrimp, "walking stick" insects, psychid moths, and some beetles.
Flatworms and earthworms are representatives of polyploids among the hermaphroditic
animals. Very few bisexually reproducing animals are polyploids. These
include a nematode parasite of horses, several species of starfish and
sea urchins, and the golden hamster. But the overall rarity of polyploids
in animals makes them the exceptions rather than the rule.
Muller has suggested that the reason why polyploidy plays a greater
role in plant evolution is that the sexes are usually separate in animals
whereas plants are often hermaphroditic. Random segregation of several
pairs of sex chromosomes in a polyploid animal would result in sterile
combination whereas polyploids with even pairs of the haploid genomes (tetraploids,
hexaploids, octaploids, etc.) in plants are perfectly fertile. For example,
a male tetraploid animal of sex chromosome constitution XXYY, and
a female tetraploid XXXX would produce gametes XY and XX,
respectively. The union of these gametes gives rise to a zygote XXXY,
and in some species this is neither completely male nor female; therefore,
it is sterile. This explanation seems to be supported by observations and
is widely accepted. Thus we can see that the above mechanism for speciation
in plants is insufficient to explain speciation in animals.
Mayr (10) maintained that by postulating different
stages a population has to go through during speciation and finding natural
populations in each of these stages, the slow past events of speciation
could be reconstructed and "proved." This approach was taken by Darwin
and other early evolutionists, but their studies were not fruitful because
of their poor definition of species. Today some of the difficulties in
defining a species have been removed by the interbreeding population
concept
(see
I.1.2),
but this concept is not always easily applicable to natural populations.
Therefore, species distinction is often arbitrary. Since there is more
than one proposed mechanism to account for speciation, the attempt to categorize
natural populations according to one's presupposition may at [205] most
yield circumstantial evidence of a highly inferential nature.

Figure 3.12. Evolution of bread wheat (Triticum aestivum)
by allopolyploidy.
In summary, empirical documentation of evolution above the species
level is not yet forthcoming. It can be argued that since macroevolution
happened over a long period of time, it cannot be observed empirically
in one's lifetime. Nonetheless, the phylogenetic developments in the higher
categories have to be extrapolated from the well-defined processes of microevolution.
The theory of organic evolution will be without a firm foundation if it
is divorced from the empirical documentation of microevolution.
For years Neo-Darwinists have asserted that natural selection, a mechanism
operating very nicely in microevolution, is equally applicable in the evolution
of the higher categories. However, it will be seen that this assertion
is seriously challenged, and some modern evolutionists maintain that natural
selection plays a minimal role in transpecific evolution, if at all. This
will be dealt with in I.3.3.2. The need to be anything but dogmatic in
one's assertion based on the evidence is immediately apparent.
d) Inconsistency of Molecular Biological Data With Data Supporting
Macroevolution. G. G. Simpson, a renowned taxonomist and paleontologist,
predicted a few decades ago the relationship between morphological changes
and genetic changes in evolution as follows (12): [206]
Morphological and taxonomic rates [of evolution]
have a decided, even though indirect, relationship to genetic rates. If
this were not so, their bearing on evolutionary theory would be quite different.
It has become commonplace that changes in morphology or phenotype may be
induced by factors other than changes in genotype and therefore may not
reflect the latter accurately. More recently it has been recognized that
changes in genotype may not be accompanied proportionately, or at all,
by changes in phenotype. Nevertheless, there can seldom be any doubt that
well-defined morphological changes in phenotypes of successive populations,
particularly as these occur over considerable periods of time in the fossil
record, run parallel to genetic changes in those populations. It is therefore
a proper assumption in such cases that morphological rates do reflect genetic
rates, even though they are probably not exactly proportioned to the latter.
The assumption is even more reliable for taxonomic rates because the concepts
and usages of modern taxonomy are in part genetical even when the observed
data are morphological.
As Simpson made clear, it has been held as the most reasonable assumption
by most evolutionists that the rate of morphological evolution reflects
the rate of genetic evolution. This was true until the advent of molecular
techniques and their use in the analysis of the genetic differences of
natural populations.
Lewontin delineated four required criteria for estimating genotypic
frequencies in populations in order to classify individuals into genetic
classes unambiguously (13): (1) Phenotypic differences
caused by the substitution of one allele for another at a single locus
must be detectable as an unambiguous difference between individuals. (2)
Allelic substitutions at one locus must be distinguishable in their effects
from allelic substitution at other loci. (3) All, or a very large fraction
of, allelic substitutions at a locus must be detectable and distinguishable
from each other, irrespective of the intensity or range of their physiological
effects. (4) The loci that are amenable to attack must be a random sample
of genes with respect to the amount of genetic variation that exists at
the locus. He concluded that the methods of classical genetics that try
to decipher the genotypes by examining the morphological and visible changes
in the organisms fail to fulfill these criteria.
The best solution to detect genetic differences in populations appears
to be the methods of molecular genetics. Since the sequence of nucleotides
that makes up a structural gene is translated into the primary structure
of a polypeptide chain with high fidelity (see I.2.6.2), the change
in the amino acid sequence reflects the mutation in nucleotides with a
high degree of colinearity. Therefore, the analysis of amino acid sequence
of proteins is a method that can satisfy all the requirements listed above.
A single allelic substitution is detectable unambiguously since it results
[207] in a discrete change in the phenotype — a substitution, deletion,
or addition of an amino acid. Every substitution is detectably different
except for mutation within the degenerate codes. The gene effects of different
loci cannot be confused with each other since they encode different proteins.
The conflict between the discrete phenotypic effect demanded by Mendelism
and the subtle phenotypic differences hardly detectable is resolved by
looking at the gene product directly. By equating one gene to one polypeptide,
the techniques in molecular genetics allow the examination of random samples
of genes regardless of their variabilities or mutabilities.
There are several methods developed in molecular genetics to analyze
genic differences among organisms.
(1) Method of DNA Hybridization. Since DNA carries the genetic
information, the most direct method of detecting genetic differences of
organisms is to measure the proportion of nucleotide pairs that are different
in their DNAs. This can be accomplished by making hybrid molecules from
single-stranded DNAs obtained by separating the double helices of the tested
organisms by physical means. When the single stranded DNAs from different
organisms are brought together, they will form hybrid duplex molecules
according to their degree of DNA homology.
Two different measures of DNA differences can be obtained by hybridization:
(1) the fraction of the DNA of two species that form hybrid molecules and
(2) the proportion of nucleotide pairs that are complementary to each other
in the hybrid molecules. The first measure can be obtained by selectively
isolating the hybrid molecules from the single-stranded DNA. The second
measure can be determined by monitoring the temperature at which these
hybrid molecules separate into single strands again, which is proportional
to the degree of nucleotide complementarity.
Since DNA hybridization is time consuming and the information obtained
is too crude to be related to a single gene, it is largely used for preliminary
analysis of genic differences among populations.
(2) Immunological Techniques. Immunological techniques depend
on the specificities of the antigen-antibody reaction. Antibodies are proteins
produced in vertebrates when they are exposed to foreign substances called
antigens. The specificity of the immunological reaction can also be visualized
by the lock and key theory (see Figure 2.50). The antibodies
serve as a lock and the antigen the key. Most antigens are proteins. A
single amino acid substitution in an antigen, reflected by the change in
the configuration of the key, prevents it from fitting the lock as tightly.
[208]
Immunological comparisons of proteins from different species can be
done easily. For example, blood albumin from a monkey is purified, and
the purified protein is injected into another mammal, a rabbit. The rabbit
will develop an antibody specifically against monkey albumin. One assesses
how close any other protein not used to immunize rabbits comes to the monkey
albumin injected into the rabbit by comparing the reaction between the
uninfected protein and the antibody with the reaction between the injected
antigen and the antibody produced. If the reactions are similar, the antigens
are similar. The degrees of dissimilarity between a protein used in an
immunization and a tested protein from another species is expressed as
an immunological distance between the species. Since the immunological
method is crude and produces indirect results, it is used only as a supplementary
method for the analysis of genetic differences.
(3) Electrophoretic Measurements. Since all amino acids contain
an acidic and a basic group, they behave as charged molecules under physiological
conditions. Some common amino acids also have polar side chains that have
a charge if ionized. Proteins formed by amino acids linked by peptide bonds
also behave as charged particles in solutions. At a given pH a protein
has a certain defined net charge determined by the number and type of ionizable
groups it possesses. A technique called electrophoresis is able to separate
proteins according to the net charges of each protein at a given pH.
If an allelic change at a genetic locus results in the replacement of
an amino acid having a nonpolar side chain with one that has a polar side
chain or vice versa, the net charge of the protein will be altered. This
can be detected when the protein is placed in an electric field, for it
will migrate differently. For example, a single-step change in the codon
AAC to AAA results in the substitution of the positively charged lysine
for the neutral asparagine. An even more drastic single-step change is
from AAG to GAG, which results in the substitution of a negatively charged
glutamic acid for the positively charged lysine.
The apparatus of a typical gel electrophoresis experiment is depicted
in Figure 3.13. It consists essentially of a slab of some jellylike material
(starch, agar, or a synthetic polymer) whose two ends are in contact with
the opposite poles of an electric potential. Material for electrophoresis
is introduced into the wells at one end of the gel. Any charged molecules
will move down the gel according to the force of attraction exerted by
the electric field. The gel is surrounded by a cooling jacket to prevent
overheating that may disrupt the three-dimensional structure of the proteins
and thus adversely affect its mobility in the electric field. The proteins
to [209] be analyzed are extracted from tissue of an individual organism,
subjected to several crude steps of a protein-purification scheme, and
then applied to the electrophoretic chamber. The speed of migration (mobility)
of each protein band will depend on its net charge and to a lesser degree
its molecular size. After the proteins have migrated across the gel, the
electrophoresis is stopped, and the proteins are stained and visualized.
Due to the relative ease of operation and the sensitivity of this method,
it has been widely used to detect polymorphic forms of protein extracted
from various individuals within a natural population. The identification
of widespread protein polymorphism is in part the evidence that led to
the formulation of the theory of neutral mutation. The occurrence of this
phenomenon is poorly explained by the mechanism of natural selection. This
will be discussed in more detail in the next section.
(4) Amino Acid Sequencing of Proteins. Amino acid sequencing
of proteins is by far the most accurate method of estimating genetic differences
among organisms. Proteins are extracted from tissues of animals or from
whole organisms and exhaustively purified to remove contaminating proteins.
The isolated protein is then subjected to amino acid sequencing. First,
if the protein has quaternary structure, the individual polypeptide chains
are separated and purified. Second, a sample of each polypeptide chain
is subjected to total hydrolysis, and its amino acid composition is determined.
Third, each polypeptide is broken into small peptide fragments by chemicals
(some enzymes) that attack only specific regions of the polypeptide. Fourth,
the amino acid sequence of each fragment is determined.
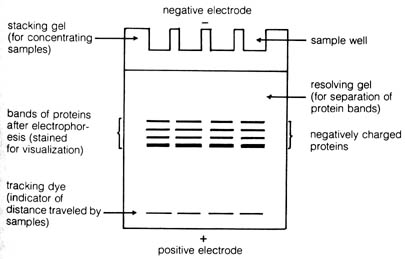
Figure 3.13. Diagram of a vertical slab gel-electrophoresis
apparatus. [210]
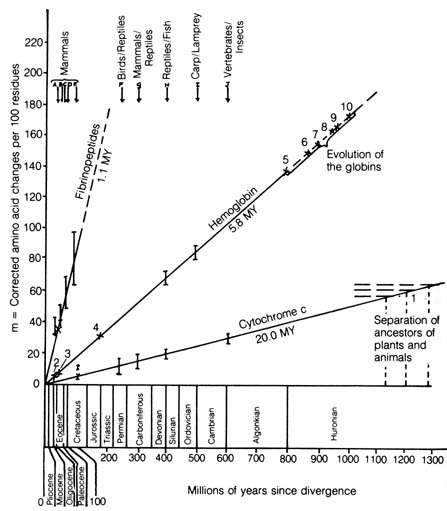
Figure 3.14. Rates of amino acid substitution in the fibrinopeptides,
hemoglobin, and cytochrome c. Comparisons for which no adequate
time coordinate is available are indicated by numbered crosses. Point 1
represents a date of 1200 ± 75 MY (million years) for the separation
of plants and animals, based on a linear extrapolation of the cytochrome
c
curve. Points 2-10 refer to events in the evolution of the globin family.
The  /
/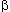 separation is at point 3,
separation is at point 3, 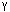 /
/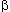 is at 4, and
is at 4, and 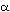 /
/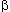 is at 500 MY (carp/lamprey). Reproduced, with permission, from Nei, M.
Molecular population genetics and evolution. New York: Elsevier &
N. Holland; 1975.
is at 500 MY (carp/lamprey). Reproduced, with permission, from Nei, M.
Molecular population genetics and evolution. New York: Elsevier &
N. Holland; 1975.
Fifth, the amino acid sequence of each polypeptide is determined
by analyzing the relationship of fragment overlap. Technological advances
have enabled the procedures to be automated in a sophisticated apparatus
known as an amino acid analyzer. [211]
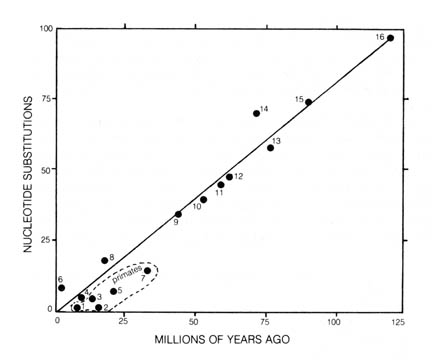 Figure 3.15. Assumed phylogeny of the species for which sequences
were examined. The order of branching was assumed. The nodes depicting
speciation are placed on the abscissa according to the maximum likelihood
solution for the number of nucleotides substituted in all seven proteins.
The upper scale does not give the true times of divergence but rather the
estimated times of divergence if one placed the marsupial-placental divergence
at 120 x 106 years ago and time was directly proportional to
the nucleotide substitution scale. Numbers are by increasing abscissal
value. Reproduced, with permission, from Fitch, W. M.; Langley, C. H. Federation
proceedings. 35:2093; 1976.
Figure 3.15. Assumed phylogeny of the species for which sequences
were examined. The order of branching was assumed. The nodes depicting
speciation are placed on the abscissa according to the maximum likelihood
solution for the number of nucleotides substituted in all seven proteins.
The upper scale does not give the true times of divergence but rather the
estimated times of divergence if one placed the marsupial-placental divergence
at 120 x 106 years ago and time was directly proportional to
the nucleotide substitution scale. Numbers are by increasing abscissal
value. Reproduced, with permission, from Fitch, W. M.; Langley, C. H. Federation
proceedings. 35:2093; 1976.
The results of amino acid sequencing are indisputably accurate,
allowing one to determine relative similarity of sequences of proteins
derived from different sources. Amino acid sequences have been studied
extensively for the proteins cytochrome c, hemoglobin, and fibrinopeptides.
This information has been used in studies of phylogeny. The results pose
some serious problems for evolutionists who advocate the mechanism of natural
selection.
When studying phylogeny using sequences of protein, the number of amino
acid substitutions that differentiates the same protein extracted from
two different organisms is plotted (14). This is plotted
against the time that the lineages of the two organisms presumably diverged
in the geological record. The results obtained are straight lines in the
case of [212] each protein (14) (Figure 3.14). The number
in millions of years (as obtained from the fossil record) listed parallel
to and beneath each line represents the estimated time necessary for a
single amino acid substitution to take place per 100 residues in the polypeptide
chains. Thus, the rate of protein evolution is roughly constant over most
of evolutionary time.
A comparison of the amino acid substitution rate of hemoglobins verses
cytochrome c using sources such as the human, rabbit, snapping turtle,
tuna, and rattlesnake shows them to be significantly different (15). Nevertheless,
when the total nucleotide substitutions as calculated from the observed
amino acid substitutions in seven proteins (cytochrome c, fibrinopeptides
A and B, hemoglobin 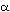 and
and 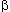 , myoglobin, and
insulin C’ peptide) have been calculated by comparisons between pairs of
mammalian species (Figure 3.15) and plotted against the time of presumed
divergence [213] of the ancestors of the respective species, a straight
line is obtained (Figure 3.16).
, myoglobin, and
insulin C’ peptide) have been calculated by comparisons between pairs of
mammalian species (Figure 3.15) and plotted against the time of presumed
divergence [213] of the ancestors of the respective species, a straight
line is obtained (Figure 3.16).
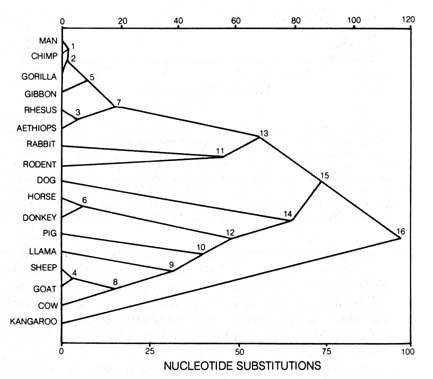
Figure 3.16. Linear relation between time elapsed and
nucleotide substitutions. Numbers on points identify the nodes of Figure
3.15 that have been plotted according to the number of nucleotide substitutions
expected from maximum likelihood solution. The line was simply drawn through
the origin and point 16. Reprinted, with permission, from Fitch, W. M.;
Langley, C. H. Federation proceedings. 35:2093; 1976.
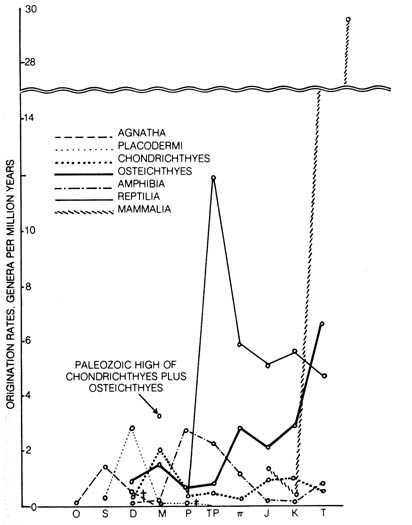
Figure 3.17. Rates of evolution in the classes of vertebrates
(except birds). The time scale runs from Ordovician (O) to Tertiary
(T). The abbreviations are as follows: S (Silurian), D
(Devonian),
M (Mississippian), P (Pennsylvanian), TP
(Permian),
TT (Triassic), J (Jurassic), K (Cretaceous).
For the time scale in years, see Table 2.8. Reprinted, with permission,
from Simpson, G. G.
The meaning of evolution. New Haven: Yale University
Press; 1949.
The presumed time of divergence of the lineages tested were constructed
from protein sequence data. This was done independent of accepted [214]
phylogenetic schemes based on geological and fossil material. Therefore,
the average rates of protein evolution over a period of time are constant
and may be used as an approximate evolutionary clock. The overall correlation
is fairly good except for the primates. They appear to have evolved at
a substantially slower rate than the average of other organisms (16).
The rates of evolution as judged by structural changes and diversification
of lines of descent, as expressed in the rate of origination of genera
per million years measured in several classes of vertebrates throughout
the geological eras, show an erratic pattern (17) (Figure
3.17). This evolution of morphological features and diversification of
descent seems to be independent of genetic change as measured by the substitutions
of nucleotides in the DNA.
An extensive comparison of 43 proteins extracted from humans and chimpanzees
correlating the electrophoretic studies and protein sequencing with the
techniques of DNA hybridization and immunological reactions were made.
It was reported that the genetic distances among species from different
genera within the same family are considerably larger than the genetic
distance between humans and chimpanzees, and they are in different families
(30). In other words, the anatomically and behaviorally distinct species
of human and chimpanzee that are classified in different zoological families
are found, according to these data, to be more closely related genetically
to each other than are several sibling species or congeneric species of
the frog, fruit fly, or mouse.
The rates of evolution at the molecular level among human and chimpanzee
lines of descent seem to be equal to each other after the presumed divergence
from a common ancestor. On the other hand, the biological evolution as
measured by the organismic change in the two lines seems to indicate that
the human has evolved much further than the chimpanzee after divergence
(Figure 3.18). This evidence also indicates that genetic changes are independent
of changes in morphological features during the course of evolution.
If the morphological and physiological features of an organism are the
result of gene expression controlled by the messages carried by DNA, an
assumption that is the working hypothesis for modern biologists, then the
apparent independence of the two levels of evolution seems to indicate
an inconsistency of molecular biological data with other data supporting
macroevolution. Although several hypotheses are postulated to try to account
for this inconsistency (see I.3.3.2), a solution that can be documented
empirically is not yet in sight. This inconsistency will likely remain
an enigma for evolutionists. [215]

Figure 3.18. Contrast between biological evolution and
molecular evolution since the divergence of the human and chimpanzee lineages
from a common ancestor. As shown on the left, zoological evidence indicates
that far more biological change has taken place in the human lineage (y)
than in the chimpanzee lineage (y >> x); this illustration
is adapted from that of Simpson. As shown on the right, both protein and
nucleic acid evidence indicate that as much change has occurred in chimpanzee
genes (w) as in human genes (z). Reproduced, with permission,
from Science, 188 (April 11): 107-116; 1975. American Association
for the Advancement of Science, Washington, D.C.
[Section 3.3 Continued]



References 3.3
1. Oparin, A. I. Genesis and evolutionary
development of life. New York: Academic Press; 1968: 29. [229]
2. Popper, K. R.
The logic of scientific
discovery. London: Hutchinson; 1959.
3. England, D.
A Christian view of origins.
Grand Rapids, MI: Baker; 1972.
4. Kerr, R. A.
Science. 210:42; 1980.
5. Hull, D. E.
Nature. 186:693; 1960.
6. Lehninger, A. L.
Biochemistry.
2nd. ed. New York: Worth; 1975: 1038.
7. Oparin, A. I.
Genesis and evolutionary
development (chapter 4).
8. Lehninger, A. L.
Biochemistry.
1048.
9. Mora, P. T.
The origins of prebiotic
systems and of their molecular matrices. Fox, S. W., ed. New York and
London: Academic Press; 1965: 39-52.
10. Mayr, E.
Animal species and evolution.
Cambridge, MA: Harvard Univ. Press; 1963: 488.
11. Grant, V.
Plant speciation.
New York: Columbia Univ. Press; 1971.
12. Simpson, G. G.
The major features
of evolution. New York: Columbia Univ. Press; 1953: 5.
13. Lewontin, R. C.
The genetic basis
of evolutionary change. New York and London: Columbia Univ. Press;
1974.
14. Nei, M.
Molecular population genetics
and evolution. Amsterdam: N. Holland; 1975: 231.
15. Ohta, T.; Kimura, M.
J. Mol. Evol.
1:18; 1971.
16. Fitch, W. M.; Langley, C. H.
Fed.
Proc. 35:2092; 1976.
17. Simpson, G. G.
The meaning of evolution.
New Haven, CT: Yale Univ. Press; 1949: 108.












